Discovering and Viewing PDB and 3D-Beacons structures
Jalview can be used to explore the 3D structures of sequences in an
alignment by following the steps below:
- Select the "3D Structure Data..." option
from a sequence's pop-up
menu to open the Structure
Chooser dialog box.
- If one or more structures exist in the PDB for the given
sequence, the Structure
Chooser dialog will open with them listed in the results
pane.
- However, if no structure was found, the Structure Chooser interface
will present options for manual association of PDB structures.
-
Since Jalview 2.11.2 you can also search the 3D-Beacons Network
for structures. The 3D-Beacons Network acts as a one-stop shop for protein
structures by combining and standardising data from several providers.
See 3D-Beacons Search for
instructions.
- Selecting PDB Structures
If one or more structures have been found from the PDB, you can select
the structures that you want to open and view by selecting them
with the mouse and keyboard.
By default, if structures were
discovered, then some will already be selected according to the
criteria shown in the drop-down menu. The default criteria is
'highest resolution', simply choose another to pick structures in
a different way.
- Viewing Cached Structures
If
previously downloaded structures are available for your
sequences, the structure chooser will automatically offer them
via the Cached Structures view. If you wish
to download new structures, select one of the PDBe selection
criteria from the drop-down menu.
- To view selected structures, click the "View"
button.
- Additional structure data will be downloaded with the
EMBL-EBI's dbfetch service
- SIFTS records will also
be downloaded for mapping UniProt protein sequence data to PDB
coordinates.
- A new structure viewer will open, or you will be
prompted to add structures to existing viewers (see below for details).
Structure Viewers in the Jalview Desktop
The
Jmol viewer has been included since Jalview
2.3. Jalview 2.8.2 included support for Chimera,
provided it is installed and can be launched by Jalview. ChimeraX and PyMOL
support is included from Jalview 2.11.2. The default
viewer can be configured in the Structure
tab in the Tools→Preferences dialog box.
Structure data imported into Jalview can also be processed to
display secondary structure and temperature factor annotation. See
the Annotation from Structure page
for more information.
Controlling where the new structures
will be shown
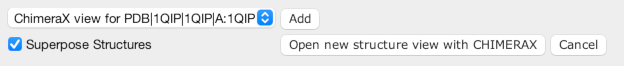
The Structure Chooser offers several options
for viewing a structure.
The Open new structure view button will open a new
structure viewer for the selected structures, but if there are views
already open, you can select which one to use, and press the Add
button. Jalview can automatically superimpose new structures based
on the linked alignments - but if this is not desirable, simple
un-tick the Superpose Structures checkbox.
Superposing structures
Jalview superposes structures
using the currently selected columns (if more than 3 columns are
selected), or the visible portions of any associated sequence
alignments. Depending on the viewer, Root Mean Squared Deviation (RMS
or RMSD) for each pair of superpositions may be output in the
structure viewer's console.
A message in the structure viewer's status bar will be shown if
not enough aligned columns were available to perform a superposition.
See the Jmol, Chimera/X
and Pymol help pages for more information
about their individual capabilities.
Retrieving sequences from the PDB
You can
retrieve sequences from the PDB using the Sequence Fetcher. The sequences
retrieved with this service are derived directly from the PDB 3D
structure data, which can be viewed in the same way above. Secondary
structure and temperature factor annotation can also be added.
Jalview will also read PDB files directly - either in PDB
format, or mmCIF. Simply load in the file
as you would an alignment file. The sequences of any protein or
nucleotide chains will be extracted from the file and viewed in the
alignment window.
Associating a large number of PDB files to
sequences in an alignment
It is often the case when working
with structure alignments that you will have a directory of PDB
files, and an alignment involving one or more of the structures. If
you drag a number of PDB files onto an alignment in the Jalview
desktop, Jalview will give you the option of associating PDB files
with sequences that have the same filename. This means, for example,
you can automatically associate PDB files with names like '1gaq.pdb'
with sequences that have an ID like '1gaq'.
Note:
This feature was added in Jalview 2.7
Note for Jalview applet users:
Due to the applet
security constraints, PDB Files can currently only be imported by
cut and paste of the PDB file text into the text box opened by the
'From File' entry of the structure menu.
Viewing the PDB Residue Numbering
Sequences which have PDB entry or PDB file associations are
annotated with sequence features from a group named with the
associated PDB accession number or file name. Each feature gives the
corresponding PDB Residue Number for each mapped residue in the
sequence. The display of these features is controlled through the "View→Sequence
Features" menu item and the Feature
Settings dialog box.
Switching between mmCIF and PDB format for
downloading files from the PDB
Jalview now employs the mmCIF format for importing 3D structure data
from flat file and EMBL-PDBe web-service, as recommended by the
wwwPDB. If you prefer (for any reason) to download data as PDB files
instead, then first close Jalview, and add the following line to
your .jalview_properties file:
PDB_DOWNLOAD_FORMAT=PDB
When this setting is configured, Jalview will only request
PDB format files from EMBL-EBI's PDBe.
mmCIF format
file support was added in Jalview 2.10.
Outstanding problem with cut'n'pasted
files in Jalview 2.6 and Jalview 2.7
Structures imported
via the cut'n'paste dialog box will not be correctly highlighted
or coloured when they are displayed in structure views, especially
if they contain more than one PDB structure. See the bug report at
http://issues.jalview.org/browse/JAL-623 for news on this problem.
Selection of sequences in the sequence panel
highlights their annotations
When sequences are selected in
the sequence panel, the corresponding annotations in the annotation
panel are highlighted. This allows the user to easily identify the
annotations associated with the selected sequence(s).
